Researchers at the Ludwig Cancer Research have surfaced a remarkable discovery in the realm of cancer research. They have identified a specific duo of genes which are involved in shaping the behavior of a particular type of immune cell within tumors.
Like a hidden code, the duo genes guide these cells in during the progression of cancer.
Ratio between CXCL9 and SPP1 or the CS ratio
The study, led by Mikaël Pittet, focused on two genes – CXCL9 and SPP1. The research showed that patients whose tumor-associated macrophages boasted elevated levels of the CXCL9 gene expression were destined for brighter clinical outcomes.
(Tumor-associated macrophages like the vigilant guards patrolling within the tumor microenvironment. They either thwart cancer’s advance or unintentionally assist its growth. Their trajectory is defined by the genetic cues they receive.)
While on the other hand, patients whose immune cells exhibited elevated levels of the SPP1 were found to be in a state that was conducive to promoting tumor growth.
Genes CXCL9 and SPP1 shape Patient Outcomes
But there’s more to discover as researchers dig into the complex system of gene activities. The research shows that how much of the CXCL9 and SPP1 genes there are plays a big part in shaping the environment around tumors.
When there’s more CXCL9 compared to SPP1, the macrophages near the tumor act against it. And this effect is seen in other cell types around the tumor too. This connection between gene amounts and fighting tumors in different cell types could lead to new and exciting treatments.
Conversely, when the CXCL9 to SPP1 ratio is low, it goes hand in hand with pro-tumor gene expression signatures across the entire tumor microenvironment (TME).
This observation shows how important these genes are in controlling gene expression. And how they act in the tumor environment. This then directly affects how the disease develops.
The Potential of CS Ratio in Tailoring Treatments
As per professor Mikaël Pittet, Ludwig Lausanne, in the future, the CS ratio could become a simple way to predict patient outcomes and guide treatment decisions.
Additionally, the study’s findings about interconnected gene expressions across different cell types offer potential targets for developing new drugs. These medications may make the tumor environment more responsive to treatments like immunotherapy.
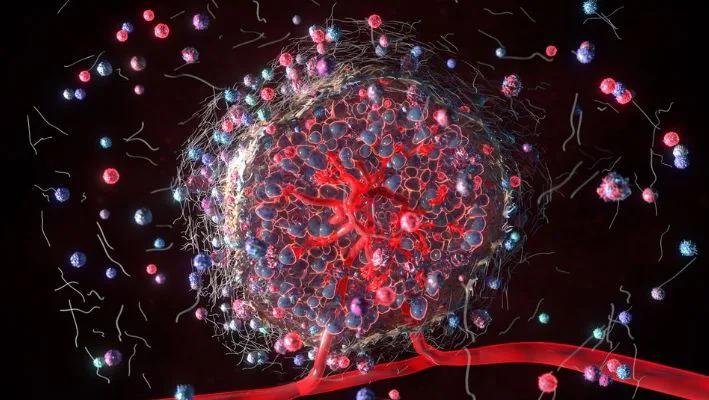
Tumor Microenvironment Cells fuel Tumor Growth
The cells surrounding the tumors are generally noncancerous cells. They are also called as tumor microenvironment (TME). And eventually, they form the foundation for tumor growth. These cells include:
- Fibroblasts – they form the tissue structures
- endothelial cells – they form blood vessels
- epithelial cells – they are responsible for lining body spaces
- some variety of immune cells – these cells can either aid or hinder tumor growth
Non-Mutating Cells in Tumor Microenvironment Key to Battling Resistance
When it comes for treatment, these cells are targeted first because they don’t mutate rapidly like cancer cells. Thus, reducing the likelihood of treatment resistance.
Therefore, Pittet and his team aimed to understand TME variations among tumors.
They studied 52 primary and metastatic tumors from 51 head and neck cancer patients. By analysing gene expressions across these tumors and correlating them with patient outcomes, they pinpointed CXCL9 and SPP1 genes as strongly connected to prognosis.
This link extended to other solid cancers too. The expression of these genes, the researchers found, better indicated whether macrophages leaned towards anti-tumor or pro-tumor effects compared to current markers.
Takeaway
In a nut shell, this study provides a remarkable leap forward in our understanding of cancer biology.
The identified genes not only hold the potential to serve as powerful predictive markers for patient outcomes, but they also open up exciting possibilities for developing novel therapeutic strategies.